PAUP*
PAUP* is a program for inference of phylogenies using parsimony, maximum-likelihood, and distance methods. It is written by David Swofford. More details are available on the PAUP* web page. You will need to download and install the command-line version of PAUP*, not the Graphical User Interface (GUI) version. If you use PAUP*, please cite it appropriately as recommended in the program's documentation.
For Windows, you should use the paup4c.exe program, not the paup4.exe program.
Note that currently PAUP* must be on a disk that is directly accessible to the computer on which you are running Mesquite.
You can conduct parsimony, likelihood, distance-based, and SVD quartets analyses from within Mesquite using Zephyr.
Parsimony Analyses
To conduct a parsimony analysis using PAUP*, choose Analysis>Tree Inference>Parsimony Tree Inference>PAUP* (Parsimony). A dialog box will appear that has four panels. The first one is critical, as that is the one in which you put the path to PAUP*.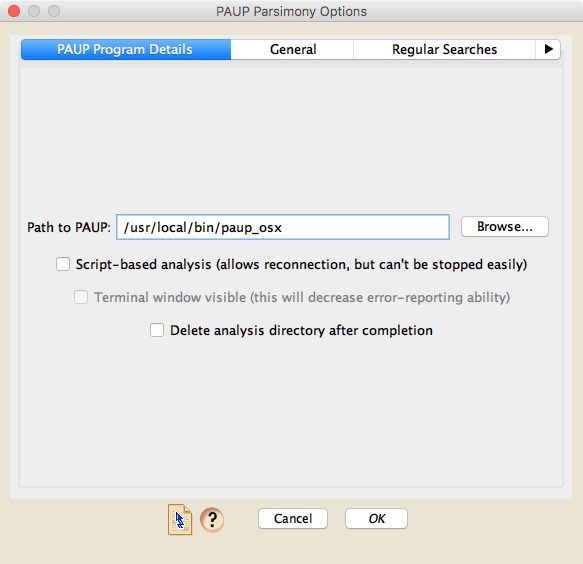
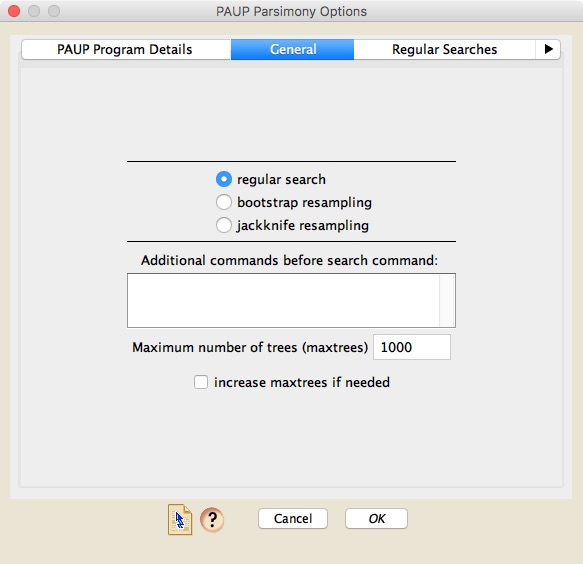
The "General" panel allows you to choose between regular searches for most parsimonious trees, or bootstrap and jackknife resampling searches. If you choose to do a regular search, then details about the search are specified in the "Regular Searches" panel; if you specify a resampling approach, then further details should be specified in the "Resampled Searches" panel.
The General panel also allows you to include PAUP* commands that will be executed before any search commands are done. You can also specify the maximum number of trees to be stored during searches, and ask PAUP* to automatically increase the maxtrees limit.
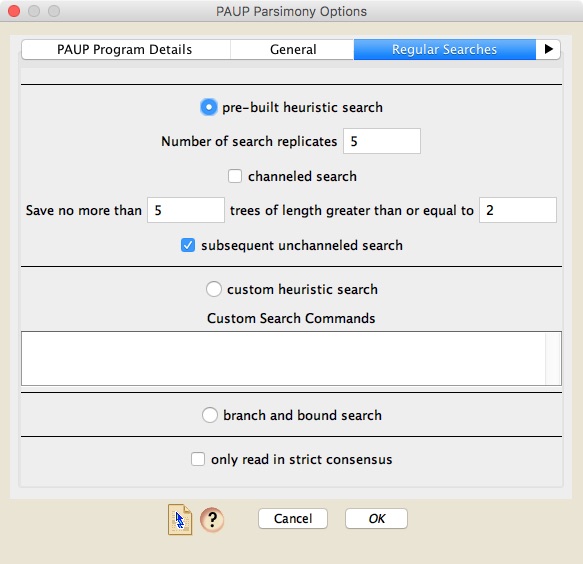
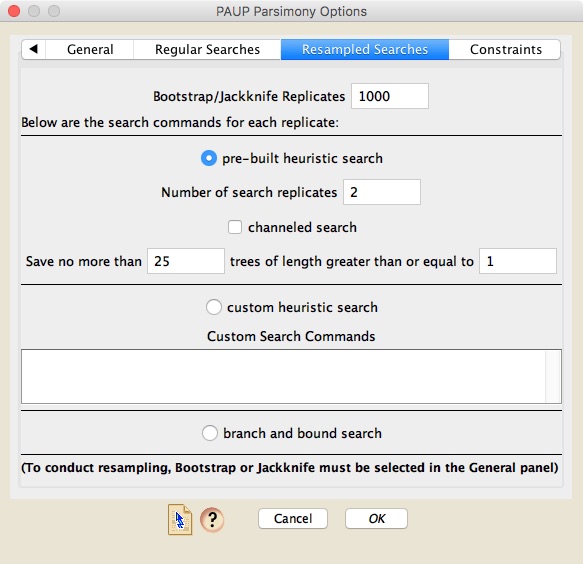
There are two ways regular searches can be conducted. Either you can ask Mesquite to design a basic search using a few of PAUP* more useful search options (this is a "pre-built search"), or you can specify your own custom search using PAUP*'s commands. If you choose a pre-built search, then you can choose how many search replicates are done, and whether the search is channeled or not. A channeled search is one in which PAUP* will restrict how many trees it stores of certain lengths, with that restriction being lifted if trees of less than a specified length are found. For example, you could ask PAUP* to store only 20 trees if the trees are longer than 100, but if it finds trees of length 99 or less, then it will store up to maxtrees. If you do ask for such a channeled search, then you can also ask PAUP* to do a subsequent unchanneled search that is a normal search, restricted only by maxtrees. [For those of you who are familiar with PAUP*'s command language, a channeled search corresponds to setting values for NCHUCK and CHUCKSCORE.]
By default, Zephyr will read in all trees produced by PAUP*. However, if you wish it to read in only the strict consensus of the trees found, then check "only read in strict consensus".
If you choose to do bootstrapping or jackknifing, then in the "Resampled Searches" panel you can choose the number of replicates, and the nature of the search for each replicate (using the same options as available for a regular search).
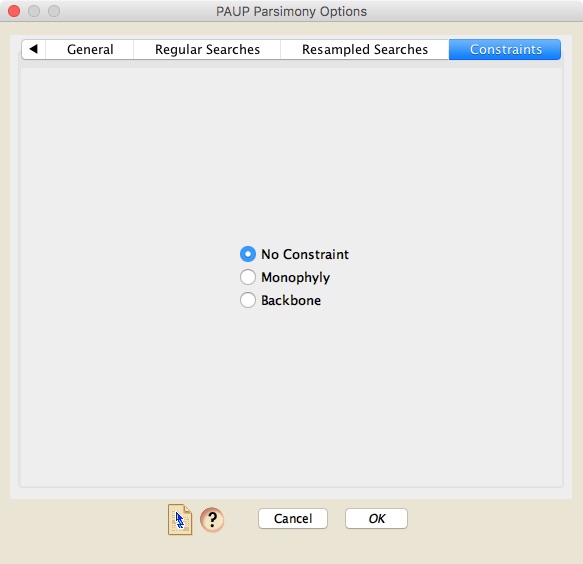
Finally, in the last panel you can specify whether a topological constraint is used or not. If you do choose to enforce a constraint, the tree to use as a constraint must be present in a Mesquite tree window in your project.
Likelihood Analyses
To conduct a parsimony analysis using PAUP*, choose Analysis>Tree Inference>Likelihood Tree Inference>PAUP* (Parsimony). A dialog box will appear that has four panels, which will be basically the same as those described under "Parsimony Analyses".Distance-based Search Analyses
To conduct a parsimony analysis using PAUP*, choose Analysis>Tree Inference>Distance Analysis>PAUP* (Parsimony). A dialog box will appear that has four panels, which will be basically the same as those described under "Parsimony Analyses".SVD Quartets
To conduct a parsimony analysis using PAUP*, choose Analysis>Tree Inference>Invariants-based Tree Inference>PAUP* (SVD Quartets) or Analysis>Tree Inference>Other Tree Inference>PAUP* (SVD Quartets). A dialog box will appear that has four panels, the first of which is like that for parsimony analyses, and the later ones simpler versions of that. There are currently no graphical buttons and fields to allow you to easily change options for an SVD analysis; instead, if you wish to alter the analysis, you will need to do this primarily in the "General" panel: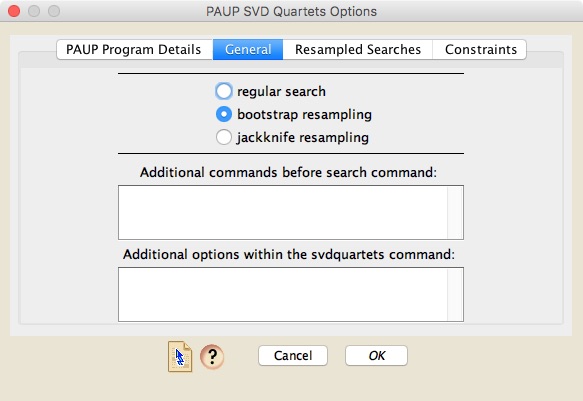
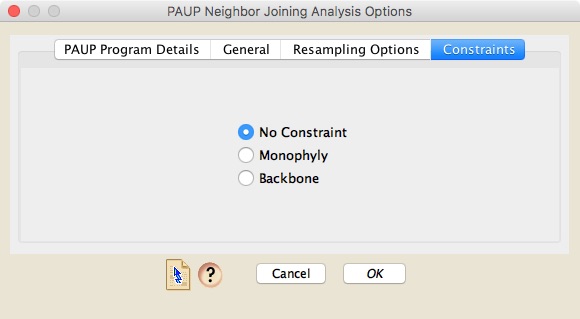
Neighbor-joining Trees
To conduct a neighbor-joining analysis using PAUP*, choose Taxa&Trees>Tree Inference>Distance Analysis>PAUP* (NJ). A dialog box will appear that has three panels. The first one is similar to the first one described under Parsimony Analysis, above.The second, "General" panel allows you to choose whether to do a simple neighbor-joining analysis (which produces just a single neighbor-joining tree), a bootstrap analyses, or a jackknife analysis.
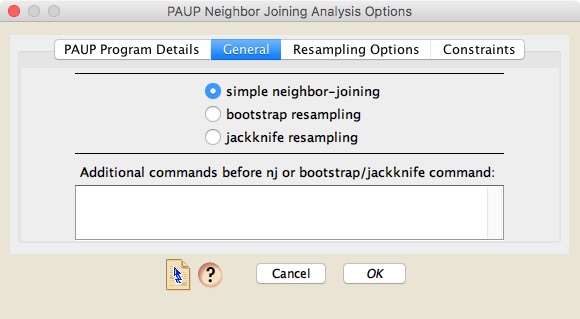
Whatever style approach you choose, you can specify additional PAUP* commands to execute before the analysis command (for example, to specify the type of distance to use).
If you choose to do a bootstrap or jackknife analysis, then you can specify the number of replicates in the "Resampling Options" panel.
Finally, the constraints panel allows you to specify whether a topological constraint is used or not. If you do choose to enforce a constraint, the tree to use as a constraint must be present in a Mesquite tree window in your project.