RAxML-NG and RAxML
RAxML-NG is a program for maximum likelihood inference of phylogenies, written by Alexey Kozlov and others; RAxML is an older program written by Alexandros Stamatakis and other code contributors. For details about RAxML-NG, go to the RAxML-NG Github page; for RAxML, go to the RAxML web page.
RAxML-NG and RAxML are distributed as apps within Mesquite for both MacOS and Linux operating systems, and thus don't require a separate installation. In addition, RAxML is distributed with Windows versions of Mesquite.
You can, if you wish, install other versions of these programs, and ask Mesquite to use those versions instead. To obtain the source code for other versions, see the RAxML-NG Github page or RAxML Github page.
If you use RAxML-NG or RAxML, please cite it appropriately as recommended in the program's documentation.
The Zephyr options available for these two programs are shown in four panels of the RAxML-NG or RAxML dialog box. The first panel is critical, as it includes basic setup details about the program; it varies depending upon whether you are running RAxML-NG or RAxML locally on your computer (the same computer that has Mesquite on it), or on an SSH server. If you have asked for the "RAxML-NG Likelihood [Local]" option, then the dialog box will look like the top image below; if you have asked for the "RAxML Likelihood [Local]" option, then the dialog box will look like the middle image:
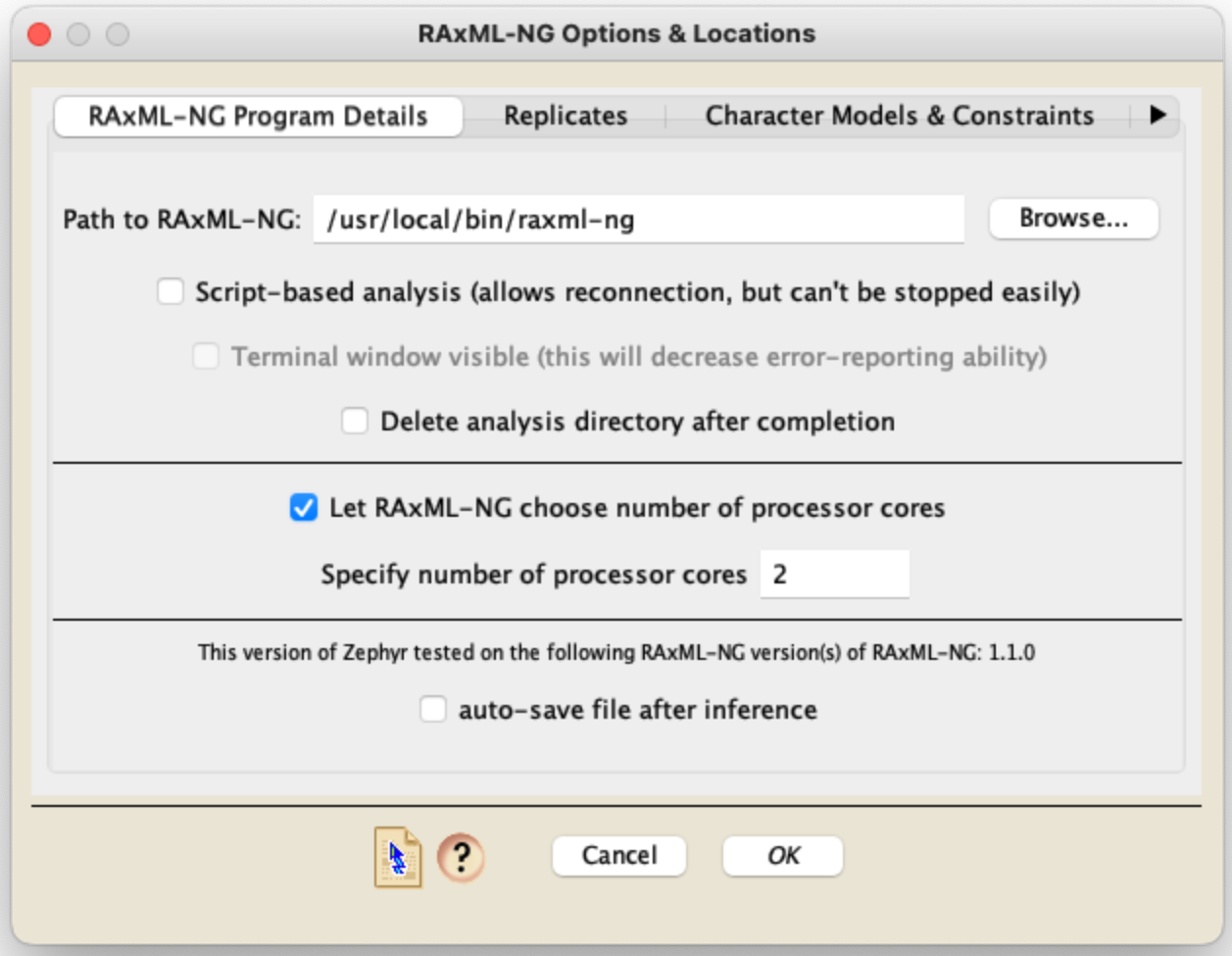
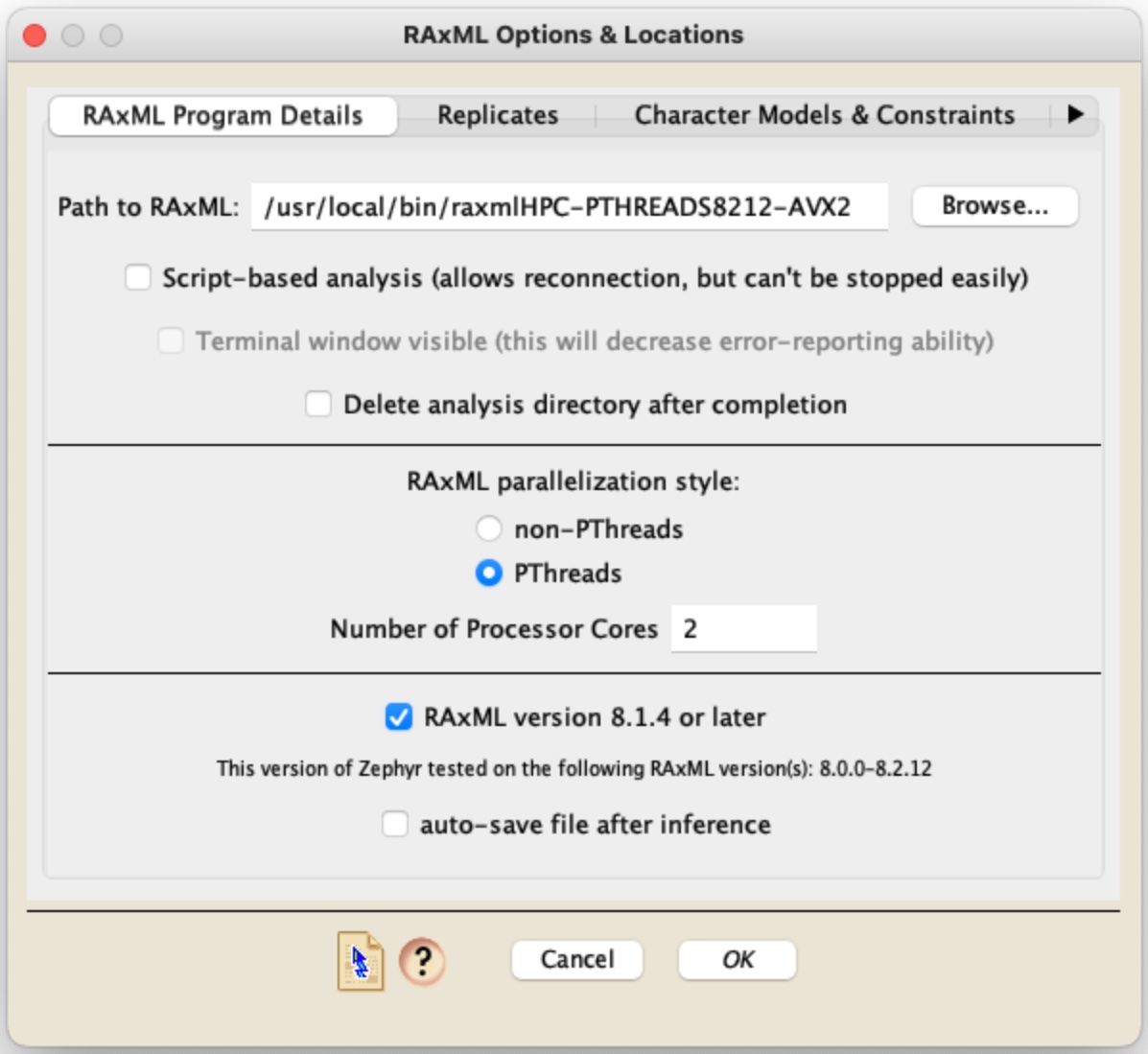
RAxML-NG or RAxML Likelihood [Local]: If RAxML-NG or RAxML are on the same computer that has Mesquite on it, then in this first panel you will need to first specify where on your computer's hard disk your copy of the software resides. The next few options (script-based, terminal, delete analysis directory) are discussed in the Local Analyses page. If you are using RAxML-NG then in this panel you will also need to choose whether to let RAxML-NG decide how many processor cores to use for the analysis; if not, then you need to set the number of cores to use. If you are using RAxML then in this panel you will also need to choose whether or not the version of RAxML you are using is a PTHREADS version or not, and the number of processors you would like RAxML to use.
Whether you are running RAxML locally or not, you have the option to automatically save the Mesquite file after the inference is completed and the phylogenetic trees moved into the file.
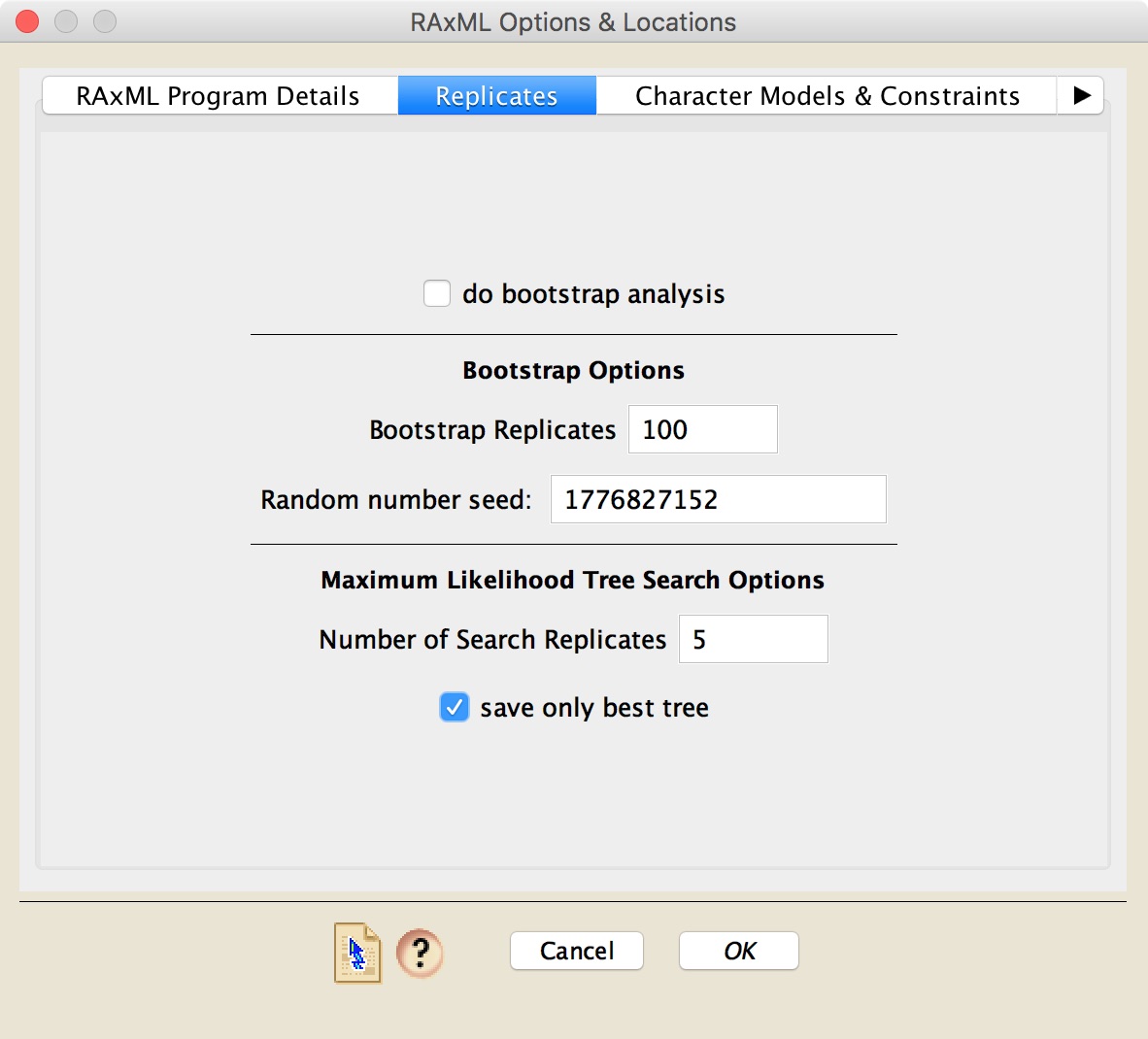
In the Replicates panel, you can choose whether to do a bootstrap analysis or a basic search of maximum likelihood trees for your data. If you choose to do a bootstrap analysis, you will need to determine the number of bootstrap replicates (or, in the case of RAxML-NG, let the program decide), as well as the random number seed. By default Zephyr will put a random number seed based upon the computer's clock, and thus each time you start a RAxML analysis the random number seed shown by default should be different.
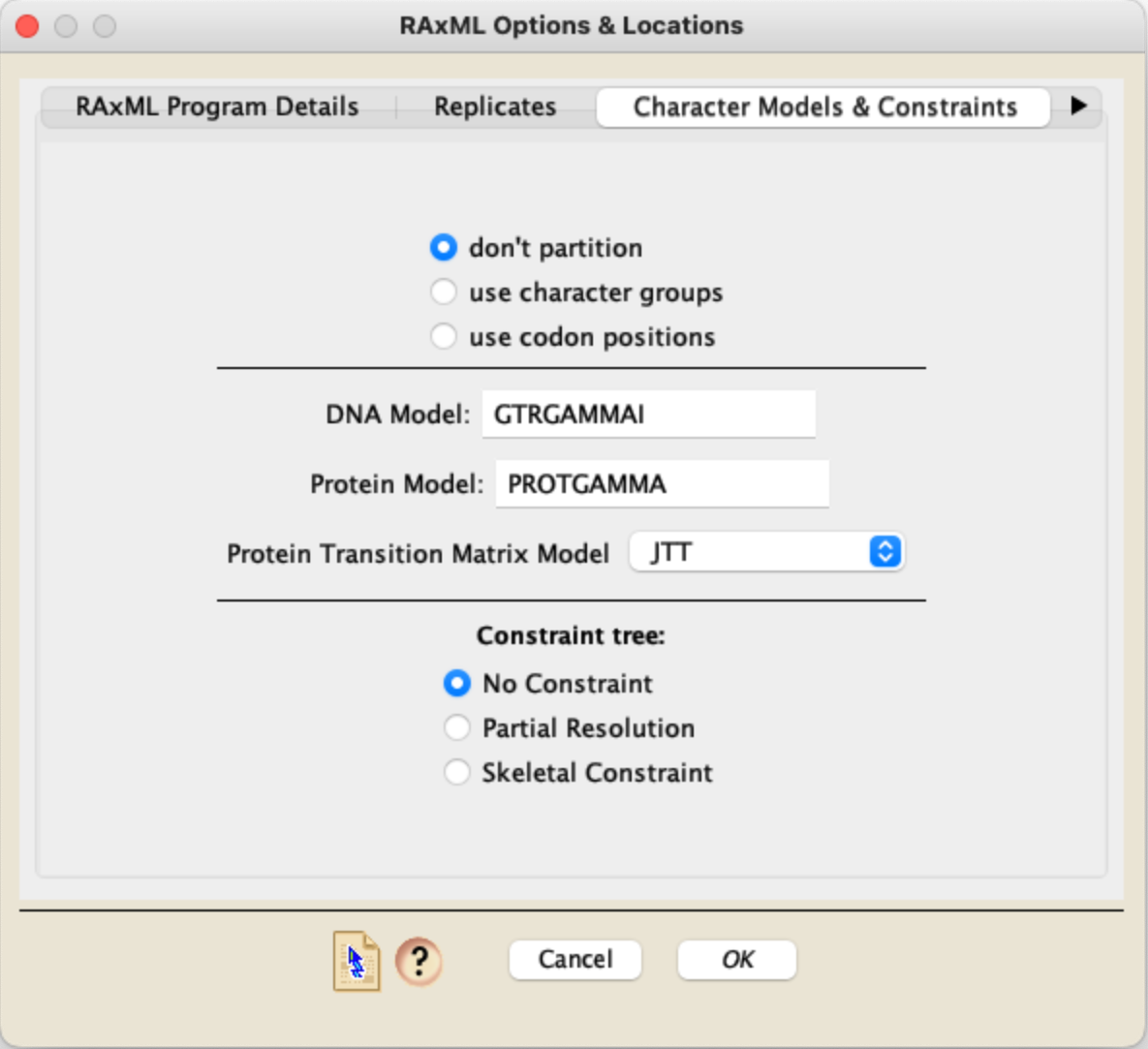
RAXML as a Character Models & Constraints panel has two options for the models to use, one for DNA data, and one for Protein data. In addition, you can choose to enforce a constraint tree. If you do choose to enforce a constraint, the tree to use as a constraint must be present in a Mesquite tree window in your project.
In contrast, RAXML-NG has a Character Models panel as follows:
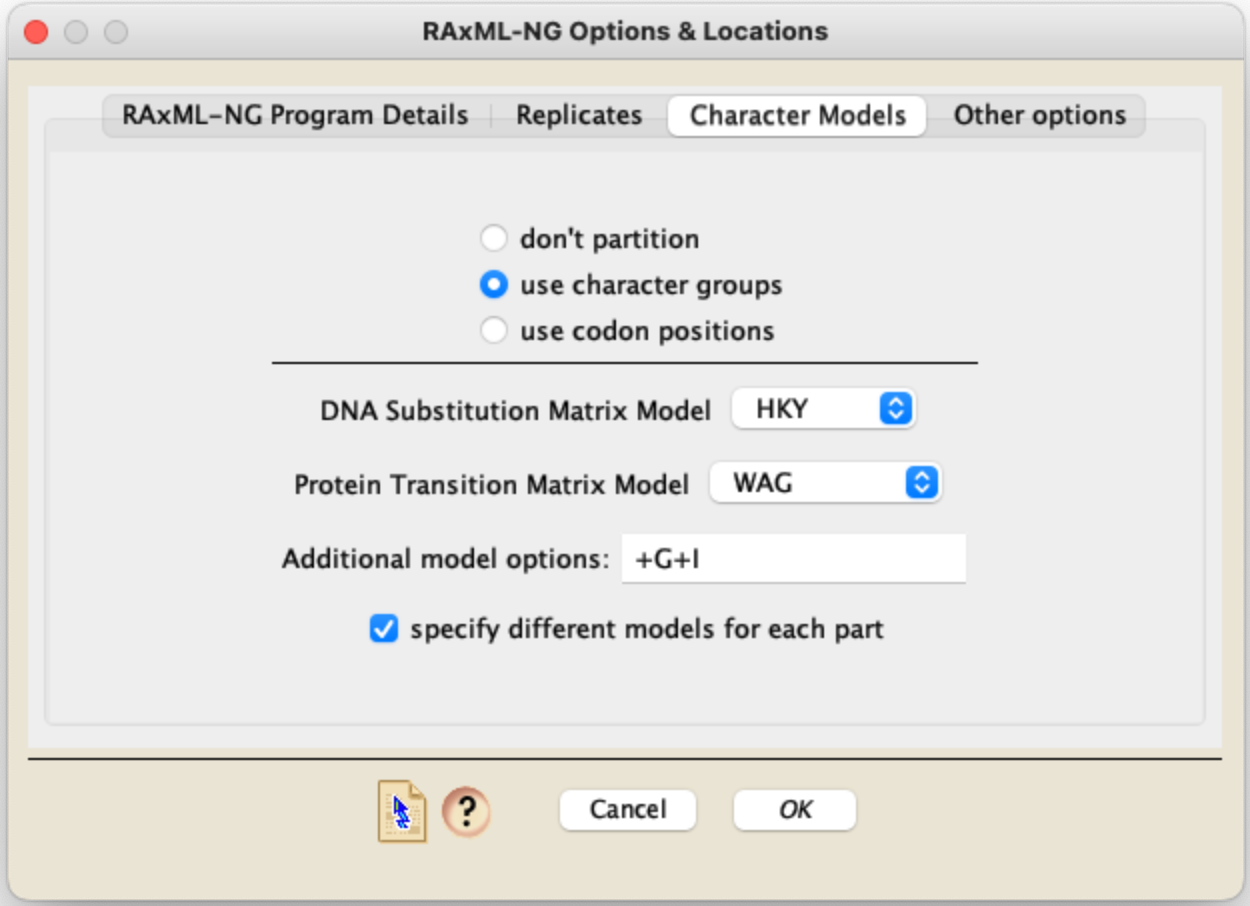
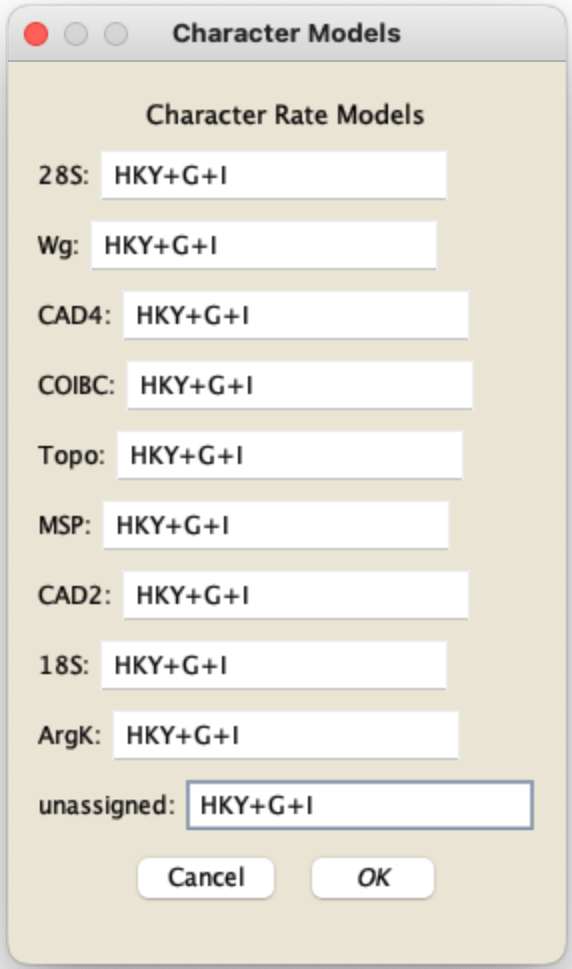
In this panel for RAXML-NG, the substitution model to be included in the RAXML-NG command is formed by taking the Matrix model (e.g., GTR for DNA in the example above), and appending the additional components from the other fields. The example shown above would yield the model specification HKY+G+I for DNA data and WAG+G+I for protein data. For additional details of the model specification, see https://github.com/amkozlov/raxml-ng/wiki/Input-data#single-model. If your data is partitioned into different character groups or codon positions and you ask those to be considered, and you check "specify different models for each part", then you will be presented with a dialog box allowing you to specify a different model for each part:
The last panel, "Other options", provides a field called "Other RAxML options" in which you can type RAxML commands yourself.
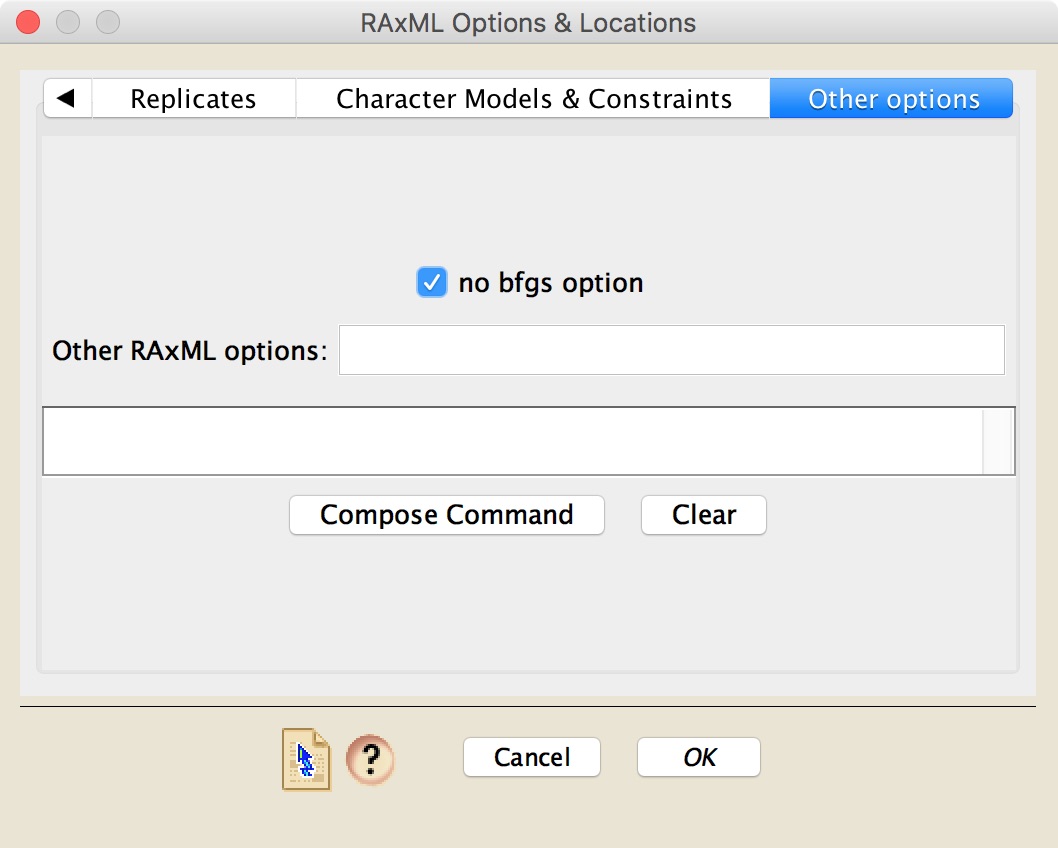
In addition, there is a "Compose Command" button that will take your options and show you the command that will be used to start RAxML and give it its options.
If you have done a non-bootstrap analysis using RAxML, then Zephyr will read in the final likelihood score of each of the RAxML trees. These scores can be seen by choosing Taxa&Trees>List of Trees>[name of RAxML tree block], and once the window listing the trees is visible, choosing Columns>Number for Tree>RAxML Score. This is the value of ln Likelihood that RAxML first reports as it is finishing each replicate. You can also see the finalized, Gamma-based score by choosing Columns>Number for Tree>RAxML Final Gamma-Based Score. Note that the latter score might not be using the same model that you used to infer the trees.
